Note
Click here to download the full example code
MNE Epochs-based pipelines#
This example shows how to use machine learning pipeline based on MNE Epochs instead of Numpy arrays. This is useful to make the most of the MNE code base and to embed EEG specific code inside sklearn pipelines.
We will compare different pipelines for P300: - Logistic regression, based on MNE Epochs - XDAWN and Logistic Regression (LR), based on MNE Epochs - XDAWN extended covariance and LR on tangent space, based on Numpy
# Authors: Sylvain Chevallier
#
# License: BSD (3-clause)
# sphinx_gallery_thumbnail_number = 2
import warnings
import matplotlib.pyplot as plt
import pandas as pd
from mne.decoding import Vectorizer
from mne.preprocessing import Xdawn
from pyriemann.estimation import XdawnCovariances
from pyriemann.tangentspace import TangentSpace
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.linear_model import LogisticRegression
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler
import moabb
from moabb.analysis.meta_analysis import ( # noqa: E501
compute_dataset_statistics,
find_significant_differences,
)
from moabb.analysis.plotting import paired_plot, summary_plot
from moabb.datasets import BNCI2014_009
from moabb.evaluations import CrossSessionEvaluation
from moabb.paradigms import P300
warnings.simplefilter(action="ignore", category=FutureWarning)
warnings.simplefilter(action="ignore", category=RuntimeWarning)
moabb.set_log_level("info")
Loading Dataset#
Load 2 subjects of BNCI 2014-009 dataset, with 3 session each
dataset = BNCI2014_009()
dataset.subject_list = dataset.subject_list[:3]
datasets = [dataset]
paradigm = P300()
Get Data (optional)#
To get access to the EEG signals downloaded from the dataset, you could
use dataset.get_data([subject_id) to obtain the EEG as MNE Epochs, stored
in a dictionary of sessions and runs.
The paradigm.get_data(dataset=dataset, subjects=[subject_id]) allows to
obtain the preprocessed EEG data, the labels and the meta information. By
default, the EEG is return as a Numpy array. With return_epochs=True, MNE
Epochs are returned.
subject_list = [1]
sessions = dataset.get_data(subject_list)
X, labels, meta = paradigm.get_data(dataset=dataset, subjects=subject_list)
epochs, labels, meta = paradigm.get_data(
dataset=dataset, subjects=subject_list, return_epochs=True
)
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
A Simple MNE Pipeline#
Using return_epochs=True in the evaluation, it is possible to design a
pipeline based on MNE Epochs input. Let’s create a simple one, that
reshape the input data from epochs, rescale the data and uses a logistic
regression to classify the data. We will need to write a basic Transformer
estimator, that complies with
sklearn convention.
This transformer will extract the data from an input Epoch, and reshapes into
2D array.
class MyVectorizer(BaseEstimator, TransformerMixin):
def __init__(self):
pass
def fit(self, X, y=None):
arr = X.get_data()
self.features_shape_ = arr.shape[1:]
return self
def transform(self, X, y=None):
arr = X.get_data()
return arr.reshape(len(arr), -1)
We will define a pipeline that is based on this new class, using a scaler and a logistic regression. This pipeline is evaluated across session using ROC-AUC metric.
mne_ppl = {}
mne_ppl["MNE LR"] = make_pipeline(
MyVectorizer(), StandardScaler(), LogisticRegression(penalty="l1", solver="liblinear")
)
mne_eval = CrossSessionEvaluation(
paradigm=paradigm,
datasets=datasets,
suffix="examples",
overwrite=True,
return_epochs=True,
)
mne_res = mne_eval.process(mne_ppl)
BNCI2014-009-CrossSession: 0%| | 0/3 [00:00<?, ?it/s]/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
BNCI2014-009-CrossSession: 33%|###3 | 1/3 [00:03<00:06, 3.39s/it]/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
BNCI2014-009-CrossSession: 67%|######6 | 2/3 [00:06<00:03, 3.03s/it]/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
BNCI2014-009-CrossSession: 100%|##########| 3/3 [00:10<00:00, 3.57s/it]
BNCI2014-009-CrossSession: 100%|##########| 3/3 [00:10<00:00, 3.46s/it]
Advanced MNE Pipeline#
In some case, the MNE pipeline should have access to the original labels from the dataset. This is the case for the XDAWN code of MNE. One could pass mne_labels to evaluation in order to keep this label. As an example, we will define a pipeline that computes an XDAWN filter, rescale, then apply a logistic regression.
mne_adv = {}
mne_adv["XDAWN LR"] = make_pipeline(
Xdawn(n_components=5, reg="ledoit_wolf", correct_overlap=False),
Vectorizer(),
StandardScaler(),
LogisticRegression(penalty="l1", solver="liblinear"),
)
adv_eval = CrossSessionEvaluation(
paradigm=paradigm,
datasets=datasets,
suffix="examples",
overwrite=True,
return_epochs=True,
mne_labels=True,
)
adv_res = mne_eval.process(mne_adv)
BNCI2014-009-CrossSession: 0%| | 0/3 [00:00<?, ?it/s]/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
BNCI2014-009-CrossSession: 33%|###3 | 1/3 [00:03<00:07, 3.66s/it]/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
BNCI2014-009-CrossSession: 67%|######6 | 2/3 [00:06<00:03, 3.13s/it]/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
BNCI2014-009-CrossSession: 100%|##########| 3/3 [00:10<00:00, 3.65s/it]
BNCI2014-009-CrossSession: 100%|##########| 3/3 [00:10<00:00, 3.56s/it]
Numpy-based Pipeline#
For the comparison, we will define a Numpy-based pipeline that relies on pyriemann to estimate XDAWN-extended covariance matrices that are projected on the tangent space and classified with a logistic regression.
sk_ppl = {}
sk_ppl["RG LR"] = make_pipeline(
XdawnCovariances(nfilter=5, estimator="lwf", xdawn_estimator="scm"),
TangentSpace(),
LogisticRegression(penalty="l1", solver="liblinear"),
)
sk_eval = CrossSessionEvaluation(
paradigm=paradigm,
datasets=datasets,
suffix="examples",
overwrite=True,
)
sk_res = sk_eval.process(sk_ppl)
BNCI2014-009-CrossSession: 0%| | 0/3 [00:00<?, ?it/s]/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
BNCI2014-009-CrossSession: 33%|###3 | 1/3 [00:05<00:10, 5.20s/it]/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
BNCI2014-009-CrossSession: 67%|######6 | 2/3 [00:10<00:05, 5.11s/it]/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 576 events (all good), 0 – 0.801 s (baseline off), ~14.5 MB, data loaded,
'Target': 96
'NonTarget': 480>
warn(f"warnEpochs {epochs}")
BNCI2014-009-CrossSession: 100%|##########| 3/3 [00:15<00:00, 5.06s/it]
BNCI2014-009-CrossSession: 100%|##########| 3/3 [00:15<00:00, 5.08s/it]
Combining Results#
Even if the results have been obtained by different evaluation processes, it is possible to combine the resulting DataFrames to analyze and plot the results.
all_res = pd.concat([mne_res, adv_res, sk_res])
We could compare the Euclidean and Riemannian performance using a paired_plot
paired_plot(all_res, "XDAWN LR", "RG LR")
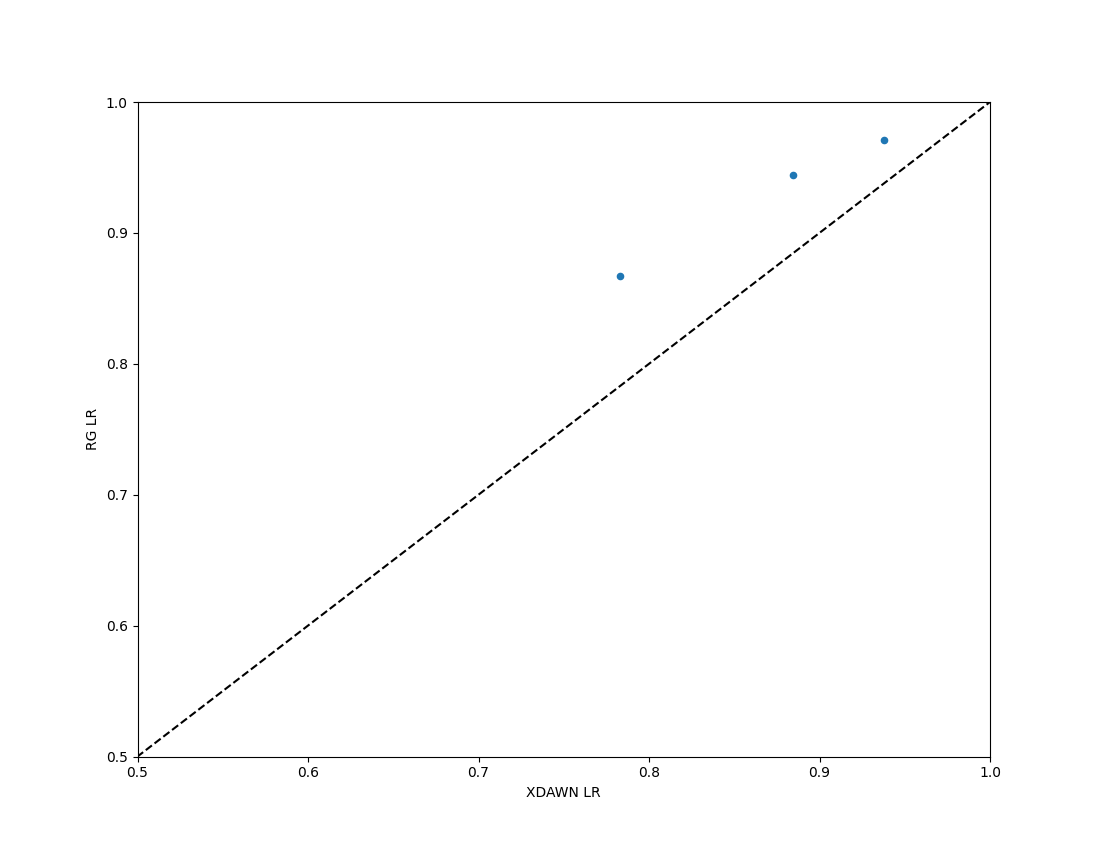
<Figure size 1100x850 with 1 Axes>
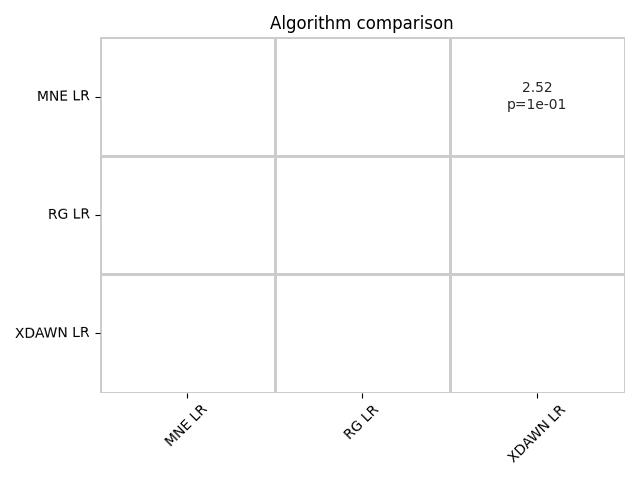
All the results could be compared and statistical analysis could highlight the differences between pipelines.
stats = compute_dataset_statistics(all_res)
P, T = find_significant_differences(stats)
summary_plot(P, T)
plt.show()
Total running time of the script: ( 0 minutes 44.768 seconds)
Estimated memory usage: 337 MB