moabb.benchmark#
- moabb.benchmark(pipelines='./pipelines/', evaluations=None, paradigms=None, results='./results/', overwrite=False, output='./benchmark/', n_jobs=-1, plot=False, contexts=None, include_datasets=None, exclude_datasets=None, n_splits=None, cache_config=None, optuna=False)[source]#
Run benchmarks for selected pipelines and datasets.
Load from saved pipeline configurations to determine associated paradigms. It is possible to include or exclude specific datasets and to choose the type of evaluation.
If particular paradigms are mentioned through select_paradigms, only the pipelines corresponding to those paradigms will be run. If no paradigms are mentioned, all pipelines will be run.
To define the include_datasets or exclude_dataset, you could start from the full dataset list, using for example the following code: > # Choose your paradigm > p = moabb.paradigms.SSVEP() > # Get the class names > print(p.datasets) > # Get the dataset code > print([d.code for d in p.datasets])
- Parameters
pipelines (str) – Folder containing the pipelines to evaluate or path to a single pipeline file.
evaluations (list of str) – If to restrict the types of evaluations to be run. By default, all 3 base types are run Can be a list of these elements [“WithinSession”, “CrossSession”, “CrossSubject”]
paradigms (list of str) – To restrict the paradigms on which evaluations should be run. Can be a list of these elements [‘LeftRightImagery’, ‘MotorImagery’, ‘FilterBankSSVEP’, ‘SSVEP’, ‘FilterBankMotorImagery’]
results (str) – Folder to store the results
overwrite (bool) – Force evaluation of cached pipelines
output (str) – Folder to store the analysis results
n_jobs (int) – Number of threads to use for running parallel jobs
n_splits (int or None, default=None) – This parameter only works for CrossSubjectEvaluation. It defines the number of splits to be done in the cross-validation. If None, the number of splits is equal to the number of subjects in the dataset.
plot (bool) – Plot results after computing
contexts (str) – File path to context.yml file that describes context parameters. If none, assumes all defaults. Must contain an entry for all paradigms described in the pipelines.
include_datasets (list of str or Dataset object) – Datasets (dataset.code or object) to include in the benchmark run. By default, all suitable datasets are included. If both include_datasets and exclude_datasets are specified, raise an error.
exclude_datasets (list of str or Dataset object) – Datasets to exclude from the benchmark run
optuna (Enable Optuna for the hyperparameter search) –
- Returns
eval_results – Results of benchmark for all considered paradigms
- Return type
DataFrame
Notes
New in version 1.1.1: Includes the possibility to use Optuna for hyperparameter search.
New in version 0.5.0: Create the function to run the benchmark
Examples using moabb.benchmark#
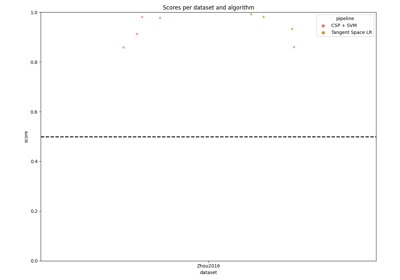
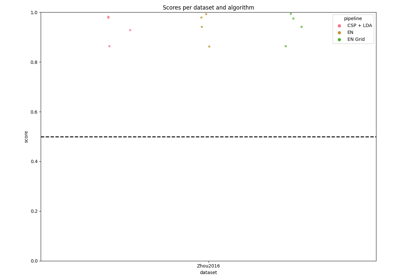
Examples of how to use MOABB to benchmark pipelines.
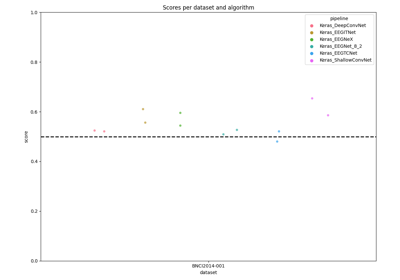
Benchmarking on MOABB with Tensorflow deep net architectures