Note
Click here to download the full example code
Tutorial 1: Simple Motor Imagery#
In this example, we will go through all the steps to make a simple BCI classification task, downloading a dataset and using a standard classifier. We choose the dataset 2a from BCI Competition IV, a motor imagery task. We will use a CSP to enhance the signal-to-noise ratio of the EEG epochs and a LDA to classify these signals.
# Authors: Pedro L. C. Rodrigues, Sylvain Chevallier
#
# https://github.com/plcrodrigues/Workshop-MOABB-BCI-Graz-2019
import warnings
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
from mne.decoding import CSP
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis as LDA
from sklearn.pipeline import make_pipeline
import moabb
from moabb.datasets import BNCI2014_001
from moabb.evaluations import WithinSessionEvaluation
from moabb.paradigms import LeftRightImagery
moabb.set_log_level("info")
warnings.filterwarnings("ignore")
Instantiating Dataset#
The first thing to do is to instantiate the dataset that we want to analyze. MOABB has a list of many different datasets, each one containing all the necessary information for describing them, such as the number of subjects, size of trials, names of classes, etc.
The dataset class has methods for:
downloading its files from some online source (e.g. Zenodo)
importing the data from the files in whatever extension they might be (like .mat, .gdf, etc.) and instantiate a Raw object from the MNE package
dataset = BNCI2014_001()
dataset.subject_list = [1, 2, 3]
Accessing EEG Recording#
As an example, we may access the EEG recording from a given session and a given run as follows:
sessions = dataset.get_data(subjects=[1])
This returns a MNE Raw object that can be manipulated. This might be enough for some users, since the pre-processing and epoching steps can be easily done via MNE. However, to conduct an assessment of several classifiers on multiple subjects, MOABB ends up being a more appropriate option.
subject = 1
session_name = "0train"
run_name = "0"
raw = sessions[subject][session_name][run_name]
Choosing a Paradigm#
Once we have instantiated a dataset, we have to choose a paradigm. This object is responsible for filtering the data, epoching it, and extracting the labels for each epoch. Note that each dataset comes with the names of the paradigms to which it might be associated. It would not make sense to process a P300 dataset with a MI paradigm object.
print(dataset.paradigm)
imagery
For the example below, we will consider the paradigm associated to left-hand/right-hand motor imagery task, but there are other options in MOABB for motor imagery, P300 or SSVEP.
We may check the list of all datasets available in MOABB for using with this paradigm (note that BNCI2014_001 is in it)
print(paradigm.datasets)
[<moabb.datasets.bnci.BNCI2014_001 object at 0x7faf5fe51cd0>, <moabb.datasets.bnci.BNCI2014_004 object at 0x7faf1de10850>, <moabb.datasets.gigadb.Cho2017 object at 0x7faf1de10310>, <moabb.datasets.mpi_mi.GrosseWentrup2009 object at 0x7faf1c145280>, <moabb.datasets.Lee2019.Lee2019_MI object at 0x7faf1dcfd250>, <moabb.datasets.liu2024.Liu2024 object at 0x7faf1dcfd790>, <moabb.datasets.physionet_mi.PhysionetMI object at 0x7faf6845d6a0>, <moabb.datasets.schirrmeister2017.Schirrmeister2017 object at 0x7faf395e9d00>, <moabb.datasets.bbci_eeg_fnirs.Shin2017A object at 0x7faf1dc75100>, <moabb.datasets.stieger2021.Stieger2021 object at 0x7faf395e9760>, <moabb.datasets.Weibo2014.Weibo2014 object at 0x7faf2f3119a0>, <moabb.datasets.Zhou2016.Zhou2016 object at 0x7faf395e9e50>]
The data from a list of subjects could be preprocessed and return as a 3D numpy array X, follow a scikit-like format with the associated labels. The meta object contains all information regarding the subject, the session and the run associated to each trial.
X, labels, meta = paradigm.get_data(dataset=dataset, subjects=[1])
Create Pipeline#
Our goal is to evaluate the performance of a given classification pipeline (or several of them) when it is applied to the epochs from the previously chosen dataset. We will consider a very simple classification pipeline in which the dimension of the epochs are reduced via a CSP step and then classified via a linear discriminant analysis.
pipeline = make_pipeline(CSP(n_components=8), LDA())
Evaluation#
To evaluate the score of this pipeline, we use the evaluation class. When instantiating it, we say which paradigm we want to consider, a list with the datasets to analyze, and whether the scores should be recalculated each time we run the evaluation or if MOABB should create a cache file.
Note that there are different ways of evaluating a classifier; in this example, we choose WithinSessionEvaluation, which consists of doing a cross-validation procedure where the training and testing partitions are from the same recording session of the dataset. We could have used CrossSessionEvaluation, which takes all but one session as training partition and the remaining one as testing partition.
evaluation = WithinSessionEvaluation(
paradigm=paradigm,
datasets=[dataset],
overwrite=True,
hdf5_path=None,
)
We obtain the results in the form of a pandas dataframe
results = evaluation.process({"csp+lda": pipeline})
BNCI2014-001-WithinSession: 0%| | 0/3 [00:00<?, ?it/s]No hdf5_path provided, models will not be saved.
No hdf5_path provided, models will not be saved.
BNCI2014-001-WithinSession: 33%|###3 | 1/3 [00:07<00:14, 7.04s/it]No hdf5_path provided, models will not be saved.
No hdf5_path provided, models will not be saved.
BNCI2014-001-WithinSession: 67%|######6 | 2/3 [00:13<00:06, 6.81s/it]No hdf5_path provided, models will not be saved.
No hdf5_path provided, models will not be saved.
BNCI2014-001-WithinSession: 100%|##########| 3/3 [00:20<00:00, 6.71s/it]
BNCI2014-001-WithinSession: 100%|##########| 3/3 [00:20<00:00, 6.76s/it]
The results are stored in locally, to avoid recomputing the results each time. It is saved in hdf5_path if defined or in ~/mne_data/results otherwise. To export the results in CSV:
results.to_csv("./results_part2-1.csv")
To load previously obtained results saved in CSV
results = pd.read_csv("./results_part2-1.csv")
Plotting Results#
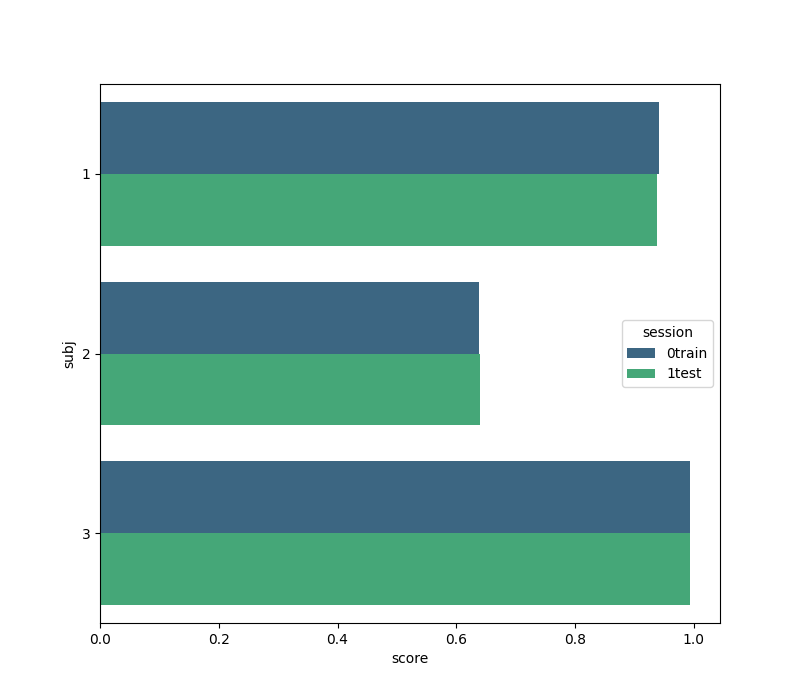
We create a figure with the seaborn package comparing the classification score for each subject on each session. Note that the ‘subject’ field from the results is given in terms of integers, but seaborn accepts only strings for its labeling. This is why we create the field ‘subj’.
Total running time of the script: ( 0 minutes 38.882 seconds)
Estimated memory usage: 640 MB