Note
Go to the end to download the full example code.
P300 Decoding
This example runs a set of machine learning algorithms on the P300 cats/dogs dataset, and compares them in terms of classification performance.
The data used is exactly the same as in the P300 load_and_visualize example.
Setup
# Some standard pythonic imports
import warnings
warnings.filterwarnings('ignore')
import os,numpy as np,pandas as pd
from collections import OrderedDict
import seaborn as sns
from matplotlib import pyplot as plt
# MNE functions
from mne import Epochs,find_events
from mne.decoding import Vectorizer
# EEG-Notebooks functions
from eegnb.analysis.analysis_utils import load_data
from eegnb.datasets import fetch_dataset
# Scikit-learn and Pyriemann ML functionalities
from sklearn.pipeline import make_pipeline
from sklearn.linear_model import LogisticRegression
from sklearn.preprocessing import StandardScaler
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis as LDA
from sklearn.model_selection import cross_val_score, StratifiedShuffleSplit
from pyriemann.estimation import ERPCovariances, XdawnCovariances, Xdawn
from pyriemann.tangentspace import TangentSpace
from pyriemann.classification import MDM
Load Data
( See the P300 load_and_visualize example for further description of this)
eegnb_data_path = os.path.join(os.path.expanduser('~/'),'.eegnb', 'data')
p300_data_path = os.path.join(eegnb_data_path, 'visual-P300', 'eegnb_examples')
# If dataset hasn't been downloaded yet, download it
if not os.path.isdir(p300_data_path):
fetch_dataset(data_dir=eegnb_data_path, experiment='visual-P300', site='eegnb_examples')
subject = 1
session = 1
raw = load_data(subject,session,
experiment='visual-P300', site='eegnb_examples', device_name='muse2016',
data_dir = eegnb_data_path)
Loading these files:
/home/runner/.eegnb/data/visual-P300/eegnb_examples/muse2016/subject0001/session001/data_2017-02-04-15_51_07.csv
/home/runner/.eegnb/data/visual-P300/eegnb_examples/muse2016/subject0001/session001/data_2017-02-04-15_58_30.csv
/home/runner/.eegnb/data/visual-P300/eegnb_examples/muse2016/subject0001/session001/data_2017-02-04-16_03_08.csv
/home/runner/.eegnb/data/visual-P300/eegnb_examples/muse2016/subject0001/session001/data_2017-02-04-15_45_13.csv
/home/runner/.eegnb/data/visual-P300/eegnb_examples/muse2016/subject0001/session001/data_2017-02-04-15_55_07.csv
/home/runner/.eegnb/data/visual-P300/eegnb_examples/muse2016/subject0001/session001/data_2017-02-04-15_47_49.csv
['TP9', 'AF7', 'AF8', 'TP10', 'Right AUX', 'stim']
['TP9', 'AF7', 'AF8', 'TP10', 'Right AUX', 'stim']
Creating RawArray with float64 data, n_channels=6, n_times=30732
Range : 0 ... 30731 = 0.000 ... 120.043 secs
Ready.
['TP9', 'AF7', 'AF8', 'TP10', 'Right AUX', 'stim']
['TP9', 'AF7', 'AF8', 'TP10', 'Right AUX', 'stim']
Creating RawArray with float64 data, n_channels=6, n_times=30732
Range : 0 ... 30731 = 0.000 ... 120.043 secs
Ready.
['TP9', 'AF7', 'AF8', 'TP10', 'Right AUX', 'stim']
['TP9', 'AF7', 'AF8', 'TP10', 'Right AUX', 'stim']
Creating RawArray with float64 data, n_channels=6, n_times=30732
Range : 0 ... 30731 = 0.000 ... 120.043 secs
Ready.
['TP9', 'AF7', 'AF8', 'TP10', 'Right AUX', 'stim']
['TP9', 'AF7', 'AF8', 'TP10', 'Right AUX', 'stim']
Creating RawArray with float64 data, n_channels=6, n_times=30732
Range : 0 ... 30731 = 0.000 ... 120.043 secs
Ready.
['TP9', 'AF7', 'AF8', 'TP10', 'Right AUX', 'stim']
['TP9', 'AF7', 'AF8', 'TP10', 'Right AUX', 'stim']
Creating RawArray with float64 data, n_channels=6, n_times=30732
Range : 0 ... 30731 = 0.000 ... 120.043 secs
Ready.
['TP9', 'AF7', 'AF8', 'TP10', 'Right AUX', 'stim']
['TP9', 'AF7', 'AF8', 'TP10', 'Right AUX', 'stim']
Creating RawArray with float64 data, n_channels=6, n_times=30732
Range : 0 ... 30731 = 0.000 ... 120.043 secs
Ready.
Filteriing
raw.filter(1,30, method='iir')
Filtering raw data in 6 contiguous segments
Setting up band-pass filter from 1 - 30 Hz
IIR filter parameters
---------------------
Butterworth bandpass zero-phase (two-pass forward and reverse) non-causal filter:
- Filter order 16 (effective, after forward-backward)
- Cutoffs at 1.00, 30.00 Hz: -6.02, -6.02 dB
Epoching
# Create an array containing the timestamps and type of each stimulus (i.e. face or house)
events = find_events(raw)
event_id = {'Non-Target': 1, 'Target': 2}
epochs = Epochs(raw, events=events, event_id=event_id,
tmin=-0.1, tmax=0.8, baseline=None, reject={'eeg': 100e-6}, preload=True, verbose=False, picks=[0,1,2,3])
print('sample drop %: ', (1 - len(epochs.events)/len(events)) * 100)
epochs
1161 events found on stim channel stim
Event IDs: [1 2]
sample drop %: 1.5503875968992276
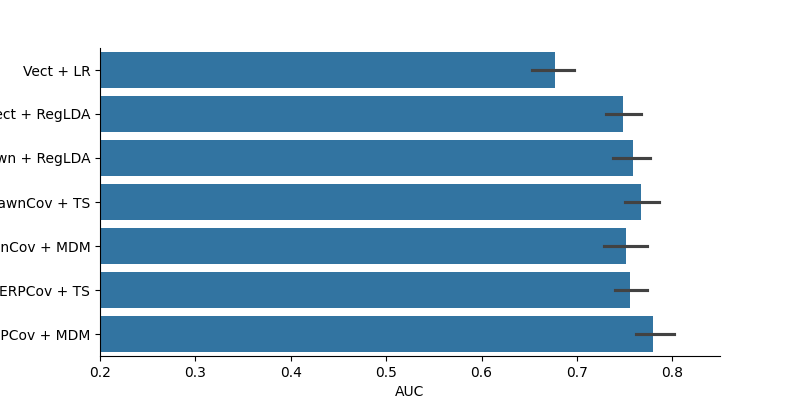
Classfication
clfs = OrderedDict()
clfs['Vect + LR'] = make_pipeline(Vectorizer(), StandardScaler(), LogisticRegression())
clfs['Vect + RegLDA'] = make_pipeline(Vectorizer(), LDA(shrinkage='auto', solver='eigen'))
clfs['Xdawn + RegLDA'] = make_pipeline(Xdawn(2, classes=[1]), Vectorizer(), LDA(shrinkage='auto', solver='eigen'))
clfs['XdawnCov + TS'] = make_pipeline(XdawnCovariances(estimator='oas'), TangentSpace(), LogisticRegression())
clfs['XdawnCov + MDM'] = make_pipeline(XdawnCovariances(estimator='oas'), MDM())
clfs['ERPCov + TS'] = make_pipeline(ERPCovariances(), TangentSpace(), LogisticRegression())
clfs['ERPCov + MDM'] = make_pipeline(ERPCovariances(), MDM())
# format data
epochs.pick_types(eeg=True)
X = epochs.get_data() * 1e6
times = epochs.times
y = epochs.events[:, -1]
# define cross validation
cv = StratifiedShuffleSplit(n_splits=10, test_size=0.25, random_state=42)
# run cross validation for each pipeline
auc = []
methods = []
for m in clfs:
res = cross_val_score(clfs[m], X, y==2, scoring='roc_auc', cv=cv, n_jobs=-1)
auc.extend(res)
methods.extend([m]*len(res))
results = pd.DataFrame(data=auc, columns=['AUC'])
results['Method'] = methods
plt.figure(figsize=(8, 4))
sns.barplot(data=results, x='AUC', y='Method')
plt.xlim(0.2, 0.85)
sns.despine()
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
Total running time of the script: (0 minutes 11.633 seconds)