Note
Go to the end to download the full example code.
Cueing Behavioural Analysis Winter 2019
Setup
# Standard Pythonic imports
import os,sys,glob,numpy as np,pandas as pd
import matplotlib.pyplot as plt
import scipy.io as sio
# EEG-Notebooks imports
from eegnb.datasets import datasets
Download the data
eegnb_data_path = os.path.join(os.path.expanduser('~/'),'.eegnb', 'data')
cueing_data_path = os.path.join(eegnb_data_path, 'visual-cueing', 'kylemathlab_dev')
# If dataset hasn't been downloaded yet, download it
if not os.path.isdir(cueing_data_path):
datasets.fetch_dataset(data_dir=eegnb_data_path, experiment='visual-cueing', site='kylemathlab_dev')
Analyze .mat behavioural data for Accuracy and RT
Load in subjects
# # Fall 2018
subs = [101, 102, 103, 104, 106, 108, 109, 110, 111, 112,
202, 203, 204, 205, 207, 208, 209, 210, 211,
301, 302, 303, 304, 305, 306, 307, 308, 309]
# 105 - no trials in one condition
# # Winter 2019
# subs = [1101, 1102, 1103, 1104, 1105, 1106, 1108, 1109, 1110,
# 1202, 1203, 1205, 1206, 1209, 1210, 1211, 1215,
# 1301, 1302, 1313,
# 1401, 1402, 1403, 1404, 1405, 1408, 1410, 1411, 1412, 1413, 1413, 1414, 1415, 1416]
# # 1107 - no csv session 1
# # 1201 - no csv session 1
# # 1304 - Muse 2
# # 1308 - Muse 2
# # 1311 - Muse 2
# # 1314 - Muse 2
# # 1407 - only session1
# Both
# Fall 2018
# subs = [101, 102, 103, 104, 106, 108, 109, 110, 111, 112,
# 202, 203, 204, 205, 207, 208, 209, 210, 211,
# 301, 302, 303, 304, 305, 306, 307, 308, 309,
# 1101, 1102, 1103, 1104, 1105, 1106, 1108, 1109, 1110,
# 1202, 1203, 1205, 1206, 1209, 1210, 1211, 1215,
# 1301, 1302, 1313,
# 1401, 1402, 1403, 1404, 1405, 1408, 1410, 1411, 1412, 1413, 1413, 1414, 1415, 1416]
Set some settings
# basic numbers
n_subs = len(subs)
n_sesh = 2
conditions = ['valid','invalid']
n_cond = len(conditions)
# cutoff trials that are too slow or fast
rt_toofast = 250
rt_tooslow = 1500
#creates arrays to save output
count_rt = np.zeros((n_subs, n_sesh, n_cond))
median_rt = np.zeros((n_subs, n_sesh, n_cond))
prop_accu = np.zeros((n_subs, n_sesh, n_cond))
Single Subject example
#select single subject
sub = subs[0]
print('Subject - ' + str(sub))
#just one session
sesh = 1
#load file
#path = './subject' + str(sub) + '/session' + str(sesh) + '/'
path = cueing_data_path + '/muse2016/subject' + str('%04.f' %sub) + '/session' + str('%03.f' %(sesh+1)) + '/'
file = [x for x in os.listdir(path) if x.endswith('.mat')][0]
output_dict = sio.loadmat(path + file)
print(path + file)
#pull out important info
output = output_dict['output']
accuracy = output[:,6]
rt = output[:,7]
validity = output[:,3]
print(accuracy,rt,validity)
# median rt on each condition
print('')
print(rt)
print(rt[validity == 0])
print(rt[(validity == 0) & (rt <= rt_tooslow)])
validRT = np.nanmedian(rt[ (validity == 1) &
(rt >= rt_toofast) &
(rt <= rt_tooslow)])
print('Valid RT = ' + str(validRT) + ' ms')
InvalidRT = np.nanmedian(rt[ (validity == 0) &
(rt >= rt_toofast) &
(rt <= rt_tooslow)])
print('Invalid RT = ' + str(InvalidRT) + ' ms')
Subject - 101
/home/runner/.eegnb/data/visual-cueing/kylemathlab_dev/muse2016/subject0101/session002/subject101_session2_behOutput_2018-11-20-16.58.50.mat
[1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1.] [435.031485 423.867152 335.010084 434.945121 297.102568 402.462939
309.984897 352.377032 315.902263 346.695745 409.051914 337.994502
412.490445 253.304399 325.551731 451.072688 362.531119 449.032591
312.999804 293.670213 327.738388 526.629063 459.253055 355.719449
352.932556 345.884407 454.775045 350.070761 313.989374 374.227764
369.010847 345.467709 334.978303 334.730519 363.791318 451.067033
301.155788 419.084514 271.027066 423.420538 342.584127 337.859108
314.203547 339.341316 467.860489 327.351041 419.173015 340.925891
318.901094 351.947652 437.583955 290.667171 319.596349 331.919014
390.521833 412.642289 305.489389 348.551979 273.162904 347.040148
339.326291 386.592227 317.4145 393.78332 426.145324 350.753921
321.549098 313.940851 379.253401 338.095756 309.040606 403.22982
368.109211] [1. 1. 0. 1. 1. 0. 0. 1. 1. 1. 1. 1. 1. 1. 1. 0. 1. 1. 1. 0. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 0. 1. 1. 1. 1. 1. 1. 1. 1. 1. 0.
0. 1. 1. 1. 1. 1. 0. 1. 1. 1. 1. 1. 1. 0. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1.]
[435.031485 423.867152 335.010084 434.945121 297.102568 402.462939
309.984897 352.377032 315.902263 346.695745 409.051914 337.994502
412.490445 253.304399 325.551731 451.072688 362.531119 449.032591
312.999804 293.670213 327.738388 526.629063 459.253055 355.719449
352.932556 345.884407 454.775045 350.070761 313.989374 374.227764
369.010847 345.467709 334.978303 334.730519 363.791318 451.067033
301.155788 419.084514 271.027066 423.420538 342.584127 337.859108
314.203547 339.341316 467.860489 327.351041 419.173015 340.925891
318.901094 351.947652 437.583955 290.667171 319.596349 331.919014
390.521833 412.642289 305.489389 348.551979 273.162904 347.040148
339.326291 386.592227 317.4145 393.78332 426.145324 350.753921
321.549098 313.940851 379.253401 338.095756 309.040606 403.22982
368.109211]
[335.010084 402.462939 309.984897 451.072688 293.670213 419.084514
340.925891 318.901094 390.521833 386.592227]
[335.010084 402.462939 309.984897 451.072688 293.670213 419.084514
340.925891 318.901094 390.521833 386.592227]
Valid RT = 347.04014799990546 ms
Invalid RT = 363.75905900013095 ms
Loop through subjects
for isub, sub in enumerate(subs):
print('Subject - ' + str(sub))
for sesh in range(n_sesh):
# get the path and file name and load data
#path = './subject' + str(sub) + '/session' + str(sesh+1) + '/'
path = cueing_data_path + '/muse2016/subject' + str('%04.f' %sub) + '/session' + str('%03.f' %(sesh+1)) + '/'
file = [x for x in os.listdir(path) if x.endswith('.mat')][0]
output_dict = sio.loadmat(path + file)
# pull out important stuff
output = output_dict['output']
accuracy = output[:,6]
rt = output[:,7]
validity = output[:,3]
# median rt on each condition
median_rt[isub,sesh,:] = [ np.nanmedian(rt[ (validity == 1) & (rt >= rt_toofast) & (rt <= rt_tooslow)]),
np.nanmedian(rt[ (validity == 0) & (rt >= rt_toofast) & (rt <= rt_tooslow)]) ]
# proportion accurate (number accurate / count)
prop_accu[isub,sesh,:] = [ np.sum(accuracy[(validity == 1) & (rt >= rt_toofast) & (rt <= rt_tooslow)]) /
np.sum((validity == 1) & (rt >= rt_toofast) & (rt <= rt_tooslow)),
np.sum(accuracy[(validity == 0) & (rt >= rt_toofast) & (rt <= rt_tooslow)]) /
np.sum((validity == 0) & (rt >= rt_toofast) & (rt <= rt_tooslow)) ]
Subject - 101
Subject - 102
Subject - 103
Subject - 104
Subject - 106
Subject - 108
Subject - 109
Subject - 110
Subject - 111
Subject - 112
Subject - 202
Subject - 203
Subject - 204
Subject - 205
Subject - 207
Subject - 208
Subject - 209
Subject - 210
Subject - 211
Subject - 301
Subject - 302
Subject - 303
Subject - 304
Subject - 305
Subject - 306
Subject - 307
Subject - 308
Subject - 309
Average over sessions and print data
# Summary stats and collapse sessions
Out_median_RT = np.squeeze(np.nanmean(median_rt,axis=1))
Out_prop_accu = np.squeeze(np.nanmean(prop_accu,axis=1))
print('Median RT')
print(Out_median_RT)
print('Proportion Accurate')
print(Out_prop_accu)
Median RT
[[361.7079635 364.45366275]
[547.10713075 611.45526175]
[600.073256 587.019756 ]
[535.04144725 533.250092 ]
[419.24263125 439.1580675 ]
[456.3330375 631.1320265 ]
[446.95026625 556.038382 ]
[484.0763345 478.4887 ]
[443.5412585 472.331497 ]
[436.66653125 501.3115865 ]
[495.63977675 587.358365 ]
[520.97468575 973.144689 ]
[395.5098235 370.232933 ]
[489.59985525 560.08098575]
[455.6651645 500.22833625]
[662.9326765 643.0934405 ]
[488.56224425 474.7125945 ]
[482.1316255 543.0173155 ]
[713.4103845 894.0804795 ]
[485.48053725 533.6649235 ]
[519.5475525 505.339771 ]
[488.45884825 500.39610975]
[527.5814465 533.4898155 ]
[477.23867575 446.72875575]
[484.94210575 578.6717025 ]
[681.7458885 804.780758 ]
[419.736862 485.569911 ]
[435.42846425 437.12191 ]]
Proportion Accurate
[[0.98305085 0.96428571]
[1. 1. ]
[0.91680961 0.88974359]
[0.97767857 0.96428571]
[0.9822995 1. ]
[0.95403439 0.6 ]
[0.92346939 0.97727273]
[0.99019608 0.97058824]
[0.97395994 1. ]
[0.68275862 0.83333333]
[0.89914021 0.89732143]
[0.92080745 0.73333333]
[0.94716042 0.91666667]
[0.95755518 1. ]
[0.95959184 0.94949495]
[0.88296296 0.94117647]
[0.90046296 0.92857143]
[0.96551724 0.875 ]
[0.91171329 0.92857143]
[0.98333333 0.9 ]
[0.95535714 0.96153846]
[0.94187987 0.95833333]
[0.96479592 0.94736842]
[1. 0.90833333]
[0.94186047 0.8 ]
[1. 1. ]
[0.97222222 0.97222222]
[0.95718391 1. ]]
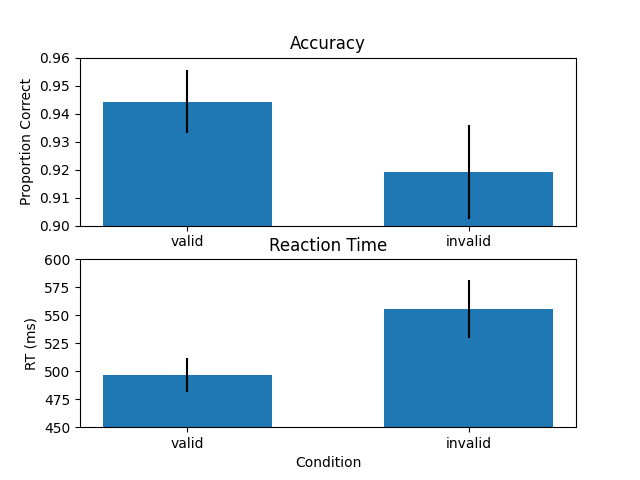
Plot barplot of results
# bar plot results
plt.figure()
# Accuracy
ax = plt.subplot(211)
plt.bar([0,1], np.nanmean(Out_prop_accu,axis=0), 0.6, yerr = np.nanstd(Out_prop_accu,axis=0)/np.sqrt(n_subs))
plt.ylim(.9,.96)
plt.title('Accuracy')
plt.ylabel('Proportion Correct')
ax.set_xticks([0,1])
ax.set_xticklabels(conditions)
# RT
ax = plt.subplot(212)
plt.bar([0,1], np.nanmean(Out_median_RT,axis=0), 0.6, yerr = np.nanstd(Out_median_RT,axis=0)/np.sqrt(n_subs))
plt.ylim(450,600)
plt.title('Reaction Time')
plt.ylabel('RT (ms)')
plt.xlabel('Condition')
ax.set_xticks([0,1])
ax.set_xticklabels(conditions)
plt.show()
Output spreadsheet
## CSV output
column_dict = {'Participant':subs,
'AccValid':Out_prop_accu[:,0],
'AccInvalid':Out_prop_accu[:,1],
'RTValid':Out_median_RT[:,0],
'RTInvalid':Out_median_RT[:,1] }
df = pd.DataFrame(column_dict)
print(df)
df.to_csv('375CueingBehPy.csv',index=False)
Participant AccValid AccInvalid RTValid RTInvalid
0 101 0.983051 0.964286 361.707963 364.453663
1 102 1.000000 1.000000 547.107131 611.455262
2 103 0.916810 0.889744 600.073256 587.019756
3 104 0.977679 0.964286 535.041447 533.250092
4 106 0.982299 1.000000 419.242631 439.158067
5 108 0.954034 0.600000 456.333038 631.132027
6 109 0.923469 0.977273 446.950266 556.038382
7 110 0.990196 0.970588 484.076334 478.488700
8 111 0.973960 1.000000 443.541259 472.331497
9 112 0.682759 0.833333 436.666531 501.311586
10 202 0.899140 0.897321 495.639777 587.358365
11 203 0.920807 0.733333 520.974686 973.144689
12 204 0.947160 0.916667 395.509823 370.232933
13 205 0.957555 1.000000 489.599855 560.080986
14 207 0.959592 0.949495 455.665165 500.228336
15 208 0.882963 0.941176 662.932677 643.093440
16 209 0.900463 0.928571 488.562244 474.712594
17 210 0.965517 0.875000 482.131626 543.017316
18 211 0.911713 0.928571 713.410385 894.080479
19 301 0.983333 0.900000 485.480537 533.664923
20 302 0.955357 0.961538 519.547552 505.339771
21 303 0.941880 0.958333 488.458848 500.396110
22 304 0.964796 0.947368 527.581447 533.489816
23 305 1.000000 0.908333 477.238676 446.728756
24 306 0.941860 0.800000 484.942106 578.671703
25 307 1.000000 1.000000 681.745889 804.780758
26 308 0.972222 0.972222 419.736862 485.569911
27 309 0.957184 1.000000 435.428464 437.121910
Total running time of the script: (0 minutes 0.157 seconds)